Uniprotkb
All uniprotkb are free cultural works licensed under a Creative Commons Attribution 4. Expert uniprotkb consists of a critical review of experimental and predicted data for each protein by a team of biologists, as well as manual verification of each protein sequence. UniProt curators extract biological information from the literature and perform numerous computational analyses, uniprotkb. Data captured from the scientific literature includes information on protein and gene names, uniprotkb, function, catalytic activity, uniprotkb, cofactors, subcellular location, protein-protein interactions and much more.
Federal government websites often end in. The site is secure. The Universal Protein Resource UniProt provides a stable, comprehensive, freely accessible, central resource on protein sequences and functional annotation. The core activities include manual curation of protein sequences assisted by computational analysis, sequence archiving, development of a user-friendly UniProt website, and the provision of additional value-added information through cross-references to other databases. For the rapid and ongoing accumulation of predicted protein sequences by high-throughput genome sequencing for numerous and increasingly diverse organisms, the expansion of large-scale proteomics e. There is a widely recognized need for a centralized repository of protein sequences with comprehensive coverage and a systematic approach to protein annotation, incorporating, integrating and standardizing data from these various sources. UniProt is the central resource for storing and interconnecting information from large and disparate sources, and the most comprehensive catalog of protein sequence and functional annotation.
Uniprotkb
The UniProt knowledgebase is a large resource of protein sequences and associated detailed annotation. The database contains over 60 million sequences, of which over half a million sequences have been curated by experts who critically review experimental and predicted data for each protein. The remainder are automatically annotated based on rule systems that rely on the expert curated knowledge. Since our last update in , we have more than doubled the number of reference proteomes to , giving a greater coverage of taxonomic diversity. We implemented a pipeline to remove redundant highly similar proteomes that were causing excessive redundancy in UniProt. The initial run of this pipeline reduced the number of sequences in UniProt by 47 million. For our users interested in the accessory proteomes, we have made available sets of pan proteome sequences that cover the diversity of sequences for each species that is found in its strains and sub-strains. To help interpretation of genomic variants, we provide tracks of detailed protein information for the major genome browsers. Protein science is entering a new era that promises to unlock many of the mysteries of the cell's inner workings. Next generation sequencing is transforming the way that we access DNA information and, as the variety of protein assays that can be linked to a DNA or RNA read-out grows, we are gaining protein information at an increasing rate. We are also gaining new insights into the mechanics of large assemblies of proteins through the incredible strides being made in electron microscopy technology. However, this wealth of molecular data will be worth little without it being available to and interpretable by the scientific community. UniProt is a long-standing collection of databases that enable scientists to navigate the vast amount of sequence and functional information available for proteins. For these entries experimental information has been extracted from the literature and organized and summarized, greatly easing scientists access to protein information. These entries are annotated by our rule based automatic annotation systems.
This systematic approach will enable proper naming and uniprotkb propagation of these sites, uniprotkb. Guidelines for investigating causality of sequence variants in human disease. I agree to the terms and conditions.
UniProt is a freely accessible database of protein sequence and functional information, many entries being derived from genome sequencing projects. It contains a large amount of information about the biological function of proteins derived from the research literature. It is maintained by the UniProt consortium, which consists of several European bioinformatics organisations and a foundation from Washington, DC , United States. Each consortium member is heavily involved in protein database maintenance and annotation. The consortium members pooled their overlapping resources and expertise, and launched UniProt in December
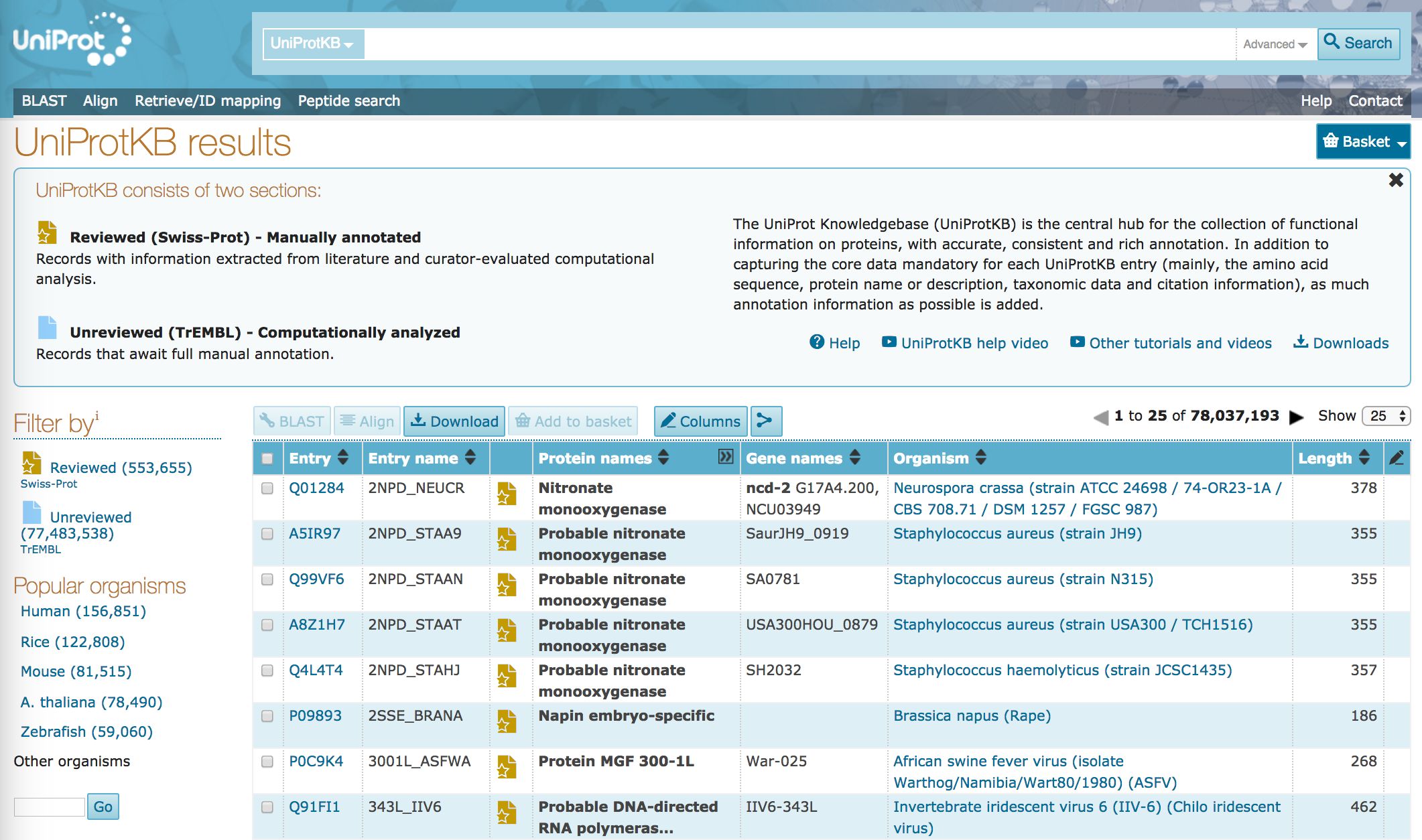
All materials are free cultural works licensed under a Creative Commons Attribution 4. Expert curation consists of a critical review of experimental and predicted data for each protein by a team of biologists, as well as manual verification of each protein sequence. UniProt curators extract biological information from the literature and perform numerous computational analyses. Data captured from the scientific literature includes information on protein and gene names, function, catalytic activity, cofactors, subcellular location, protein-protein interactions and much more. These entries are largely proteins from species for which we have no experimental data available in the scientific literature. These unreviewed records are enriched with functional annotation by systems using the protein classification tool InterPro , which classifies sequences at superfamily, family and subfamily levels, and predicts the occurrence of functional domains and important sites. Data can be searched in any of the UniProt databases using the methods described below. Once you have found an entry that interests you, click on it to open and you may then scroll down to access all the information within it, either by reading the text or visualising the information in one of the integrated viewers. You can navigate within the entry by clicking on the side-bar.
Uniprotkb
Federal government websites often end in. The site is secure. The UniProt knowledgebase is a large resource of protein sequences and associated detailed annotation.
Pierre gasly girlfriend
Rules for automatic annotation generated by database curators and computational algorithms. Spastin specifically recognizes and cuts microtubules that are polyglutamylated: severing activity by spastin increases as the number of glutamates per tubulin rises from one to eight and decreases beyond this glutamylation threshold Only the pathogenic variation in C is annotated in other public resources. Expert curation consists of a critical review of experimental and predicted data for each protein by a team of biologists, as well as manual verification of each protein sequence. Descriptions of the modified residues are structured in a machine-readable format with the use of standardized vocabularies being essential to organize knowledge for subsequent retrieval. In addition, to complement this approach, we have developed a semi-automatic pipeline for integration of high-throughput proteomics data that is distinct from expert curation and which adds PTMs from manually evaluated large-scale proteomics publications Finally we provide the UniProt Archive UniParc that provides a complete set of known sequences, including historical obsolete sequences 3. UniRef: comprehensive and non-redundant UniProt reference clusters. Smith L. The figure also shows how these variants disrupt protein structural and functional features. The full text of each paper is read, and information is extracted and added to the entry. In September , subcell. UniProt Archive UniParc is a comprehensive and non-redundant database, which contains all the protein sequences from the main, publicly available protein sequence databases. For example, Edde et al.
The aim of the UniProt Knowledgebase is to provide users with a comprehensive, high-quality and freely accessible set of protein sequences annotated with functional information. In this article, we describe significant updates that we have made over the last two years to the resource.
Please check for further notifications by email. This has made it possible to implement filters to narrow the publication list by categories that are based on the type of data a publication contains about the protein such as function, interaction, sequence, etc. Only residues that satisfy the rule criteria are ultimately propagated to entries within the PIRSF that lack an experimentally derived structure. Views 55, This allowed us to test the technical infrastructure and also to identify areas for improvements in the data representation. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. The viewer shows all features of a UniProtKB entry and includes additional features that are mapped from large-scale studies, currently for variants and proteomics data. Standard NGS annotation tools identify simple effects like translation stops and missense mutations, but more subtle effects like disruption of an enzyme active site, protein-binding site or post-translationally modified site are often not included. Citing articles via Web of Science UniProt continues to adapt its data gathering, data processing and data display to improve the availability and utility of protein information for the benefit of all. Typical examples of such sequence discrepancies may include frameshifts, erroneous gene model predictions and the presence of contaminating vector sequence or sequence of unknown origin. PLoS One. The UniRef database combines identical sequences and sub-fragments into a single UniRef entry. Search Menu. The InterPro protein families database: the classification resource after 15 years.

0 thoughts on “Uniprotkb”