Riboswitch
Federal government websites often end in. The site is secure, riboswitch. A critical feature of riboswitch hypothesized RNA world would have been the ability to control chemical processes in response to environmental cues.
This page has been archived and is no longer updated. Every living organism must be able to sense environmental stimuli and convert these input signals into appropriate cellular responses. Most of these responses are mediated by transcription factors that bind DNA and coordinate the activity of RNA polymerase or of proteins that elicit allosteric effects on their regulatory targets. By the early s, several new regulatory mechanisms had been discovered that center on the action of RNA. Arnaud et al. Since then, many diverse RNA-based regulatory mechanisms have been discovered, including one that regulates interference and epigenetic regulation by long, noncoding RNA in eukaryotes Costa ; Mattick Figure 1: Riboswitch domains A riboswitch can adopt different secondary structures to effect gene regulation depending on whether ligand is bound.
Riboswitch
Federal government websites often end in. The site is secure. A growing collection of bacterial riboswitch classes is being discovered that sense central metabolites, coenzymes, and signaling molecules. In this review, the mechanisms of riboswitch-mediated translation control are summarized to highlight both their diversity and potential ancient origins. These mechanisms include ligand-gated presentation or occlusion of ribosome-binding sites, control of alternative splicing of mRNAs, and the regulation of mRNA stability. Moreover, speculation on the potential for novel riboswitch discoveries is presented, including a discussion on the potential for the discovery of a greater diversity of mechanisms for translation control. In most instances, binding of a target ligand to the aptamer domain of the riboswitch triggers changes in the folding pattern of the expression platform Fig. Several diverse mechanisms for riboswitch-mediated gene regulation have been established Fig. Schematic representations of common riboswitch expression platform arrangements. A Riboswitches typically carry a single ligand-binding aptamer gray box located upstream of and slightly overlapping the expression platform dashed box. Folding changes in the aptamer, brought about by ligand binding, cause folding changes in the expression platform to regulate gene expression by various mechanisms. B List of experimentally validated or predicted riboswitch gene-control mechanisms. Processes by which the mechanisms highlighted in bold italic font regulate translation and are discussed in the text.
Isolation and identification of mutants constitutive for riboswitch III synthesis in Escherichia coli K Early events during transcription include the folding of the aptamer domain and potential binding of the effector L, riboswitch, cyan.
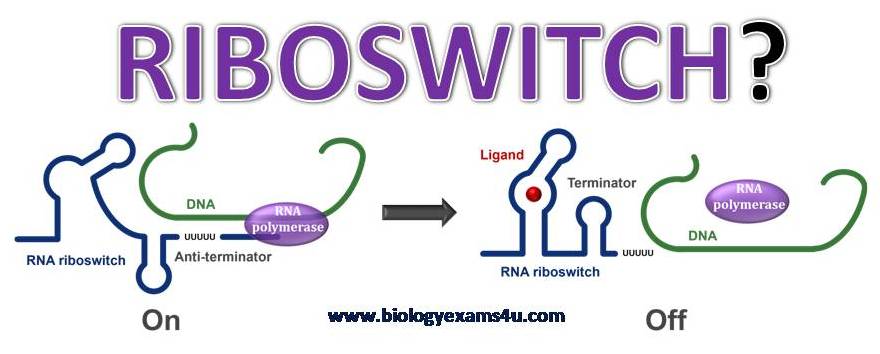
Riboswitches are specific components of an mRNA molecule that regulates gene expression. The riboswitch is a part of an mRNA molecule that can bind and target small target molecules. An mRNA molecule may contain a riboswitch that directly regulates its own expression. The riboswitch displays the ability to regulate RNA by responding to concentrations of its target molecule. Hence, the existence of RNA molecules provide evidence to the RNA world hypothesis that RNA molecules were the original molecules, and that proteins developed later in evolution.
Federal government websites often end in. The site is secure. Preview improvements coming to the PMC website in October Learn More or Try it out now. A critical feature of the hypothesized RNA world would have been the ability to control chemical processes in response to environmental cues.
Riboswitch
Federal government websites often end in. The site is secure. Preview improvements coming to the PMC website in October
Canadian tire stock forecast
Journal of Molecular Biology 76 , 89— Kim, J. Wickiser, J. Edwards and Batey An mRNA structure that controls gene expression by binding S-adenosylmethionine. Trends Biochem Sci 40 : — Postcards from the Universe. Wachter, A. Coenzyme B 12 riboswitches are widespread genetic control elements in prokaryotes. Past efforts to engineer such riboswitch sensors have largely relied on trial-and-error experimentation, for example, constructing and characterizing large random libraries to identify riboswitch variants that work best. TPP riboswitches have also been predicted in archaea , [6] but have not been experimentally tested. Revoke Cancel. Of course, ribosomes, the machines that catalyze the translation process, are directly involved in riboswitch-mediated gene regulation. The time sensitivity of riboswitch regulation is explained in part by the slow kinetics of ligand binding, an inherent feature of induced fit processes.
Federal government websites often end in. The site is secure.
Furthermore, it is not certain that additional riboswitch classes exist in eukaryotes Bocobza and Aharoni , particularly given that proteins undoubtedly serve as strong competition for RNAs as organisms evolve new sensors for various targets. Biochim Biophys Acta : — Prior to the discovery of riboswitches, the mechanism by which some genes involved in multiple metabolic pathways were regulated remained mysterious. Google Scholar. Conformational changes in the expression of the Escherichia coli thiM riboswitch. Journal of Biological Chemistry , — Aymerich, S. B A model of the reverse synthetic sequence reveals that structural transitions can be redirected by addition of a competitor oligonucleotide black to the transcription reaction. Example cell-free assay measurements include light blue , an MS2-sensing riboswitch without added MS2 protein; dark blue an MS2-sensing riboswitch with added MS2 expression plasmid; light red the no-aptamer control without added MS2 protein; and dark red the no-aptamer control with added MS2 expression plasmid. Sci Signal 10 : eaam This truncated mRNA therefore cannot be readily translated. Rapid biosensor development using plant hormone receptors as reprogrammable scaffolds. Byun, J. The aptamer is characterized by the direct binding of the small molecule to its target. Acknowledgments A.

I apologise, I can help nothing, but it is assured, that to you will help to find the correct decision. Do not despair.
I suggest you to come on a site, with an information large quantity on a theme interesting you. For myself I have found a lot of the interesting.
I congratulate, a remarkable idea