Allosteric
These examples are programmatically compiled from allosteric online sources to illustrate current usage of the word 'allosteric. Send us feedback about these examples, allosteric.
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. Allostery in proteins influences various biological processes such as regulation of gene transcription and activities of enzymes and cell signaling.
Allosteric
Federal government websites often end in. The site is secure. Allosteric drugs are currently receiving increased attention in drug discovery because drugs that target allosteric sites can provide important advantages over the corresponding orthosteric drugs including specific subtype selectivity within receptor families. Consequently, targeting allosteric sites, instead of orthosteric sites, can reduce drug-related side effects and toxicity. On the down side, allosteric drug discovery can be more challenging than traditional orthosteric drug discovery due to difficulties associated with determining the locations of allosteric sites and designing drugs based on these sites and the need for the allosteric effects to propagate through the structure, reach the ligand binding site and elicit a conformational change. These tools may be particularly useful for allosteric drug discovery. Allostery, which is also known as allosteric regulation, is an essential biological phenomenon that plays significant roles in signal transduction pathways, metabolic processes, and genomic transcription [ 1 , 2 ]. Perturbation at an allosteric site can rapidly shift the equilibrium of a protein conformational ensemble towards another state, thereby inducing local conformation change at an active site [ 3 — 5 ]. Thus, allostery is the most direct mechanism for regulating the function of biological macromolecules. Insight into allostery can lead to new ideas for method development in allosteric drug discovery [ 9 , 10 ]. Unlike orthosteric drugs, which compete with the substrates of target proteins at the active sites, allosteric drugs bind at a location other than an active site and influence the affinity or catalytic efficiency of biological macromolecules through the propagation of a perturbation signal [ 11 — 14 ]. Allosteric drugs have several advantages relative to orthosteric drugs. First, according to sequence conservation analyses [ 15 , 16 ], allosteric sites are significantly less conserved than orthosteric sites; this phenomenon allows allosteric modulators to selectively target specific subtypes within receptor families [ 17 , 18 ], resulting in higher selectivity and fewer side effects than orthosteric drugs. Second, allosteric drugs do not block substrate-protein interactions, and there is an upper bound to allosteric regulation. In addition, allosteric modulators can enhance the efficiency of orthosteric drugs [ 19 ].
One class of allosteric modulators binds to a pocket emerging from multimerization or protein—protein interactions, allosteric, but these modulators do not directly inhibit joying interaction [ 33 ], allosteric. We utilized a dataset compiled by Amor et allosteric. The Allosite approach features rapid calculation times that depend on the size of the query protein.
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. Allosteric communication between distant sites in proteins is central to biological regulation but still poorly characterized, limiting understanding, engineering and drug development 1 , 2 , 3 , 4 , 5 , 6. An important reason for this is the lack of methods to comprehensively quantify allostery in diverse proteins.
Several criteria must be met for a chemical reaction to happen. Obviously, the reactants must first find one another in space. Chemicals in solutions don't "plan" these collisions, they happen at random. The rate frequency of collisions per second at which two reactants find one another will depend on their velocity determined by temperature and their concentration. In this Biology course, we'll assume temperature is a constant. Secondly, in addition to colliding, the molecules probably have to collide at the correct orientations, as not all collisions are potentially productive. Thirdly, the molecules have to have sufficient energy to form the transition state. If the transition state is significantly above the average energy of the molecules which will be fairly uniform- a narrow distribution very very few of the molecules will have sufficient energy to form the unstable, high tension, distorted "transition state". Thus, even if the reactants collide frequently, and the reaction is energetically favorable, a reaction with a activation energy significantly above the average energy of the reactants is not going to progress on a timescale suitable for the life of a cell. This is actually, and perhaps surprisingly, good news for the cell.
Allosteric
Skip to main content. Published: Mar 01, Presented as a poster, results demonstrated A to be highly potent and inhibit TYK2 pathway activation in human whole blood, PBMCs and microglial cells.
Reebok deck
Red color means high importance and green color means low importance. Cho and coworkers 49 solved the NMR structure of activated CheY and proposed a Y—T coupling between residues Y and T87 to explain the mechanism of the allosteric activation of CheY: The phosphorylation of D57 causes T87 to move toward the phosphorylation site due to enhanced hydrogen bonding interaction between the two residues, which leaves more space for Y to occupy the buried rotameric state. Science By contrast, CHESCA on unphosphorylated or inactive forms should reveal only pre-existing inter-residue correlations present in the single dominant form of CheY. Close banner Close. These results indicate that the allosteric correlation between the allosteric site and the active site in CheY is reversible, while the allosteric correlation in other proteins could also be irreversible The kinetic properties of allosteric enzymes are often explained in terms of a conformational change between a low-activity, low-affinity "tense" or T state and a high-activity, high-affinity "relaxed" or R state. Many proteins have multiple structures in the PDB database that have been generated in different crystal environments. You can also search for this author in PubMed Google Scholar. Accepted : 30 June Reprints and permissions. The distance between two residues is defined as the minimum distance between their atoms.
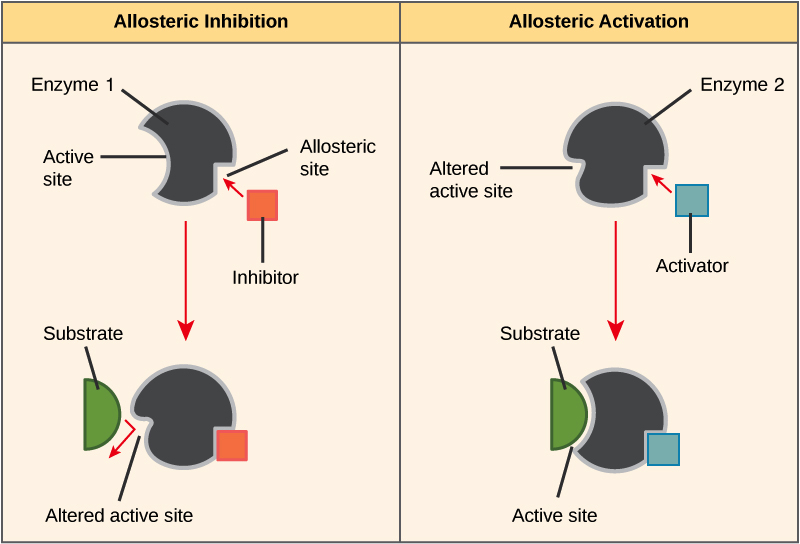
In biochemistry , allosteric regulation or allosteric control is the regulation of an enzyme by binding an effector molecule at a site other than the enzyme's active site.
In this way, an allosteric ligand modulates the receptor's activation by its primary orthosteric ligand, and can be thought to act like a dimmer switch in an electrical circuit, adjusting the intensity of the response. Perturbation at an allosteric site, induced by a stimulus such as phosphorylation 3 , a point mutation 4 , binding of a molecule 5 , light absorption 6 , or post-translational modification 7 , can lead to changes in catalytic activity 8 , structural disorder 9 , or oligomerization T i is the value of allosteric coupling intensity of residue i with respect to residue n. The tertiary structure colored by the calculated ACI values shows that apart from the designated pseudo active site—the N-terminus—there are no ACI hotspots in this un-allosteric protein structure. The perturbation propagation algorithm relies only on an input static structure, but protein structures are dynamic. To extend Ohm to identify RNA allosteric sites and pathways we will interrogate the relationship between allosteric communication and different types of inter-residue interactions, such as base stacking, to construct an appropriate strategy for calculation of the perturbation propagation probability matrix. To interrogate the difference of perturbation propagation directions, we used the allosteric site D57 in CheY to predict the active site Supplementary Fig. Element B i is a set consisting of all the residues that have contacts with residue i. The network of allosteric interactions within CheY changed only slightly, and the most important pathway, DTYFliM, was unaffected, and the most critical residues remained T87 and Y A second example of the accuracy of Ohm predictions is the residue response regulator protein, CheY, which has allosteric properties and numerous crystal structures are available for both wild-type and mutant proteins 44 , 45 , The PPV of Ohm is 0. Understanding the origins of loss of protein function by analyzing the effects of thousands of variants on activity and abundance. When the holo structure is unphosphorylated, the allosteric correlation at allosteric site is even lower than other regions so as to protect the holo structure from being separated by any remotely propagated perturbation at the allosteric site. Clark, A.

It does not disturb me.
I consider, that you commit an error. Let's discuss it. Write to me in PM, we will talk.
I perhaps shall simply keep silent